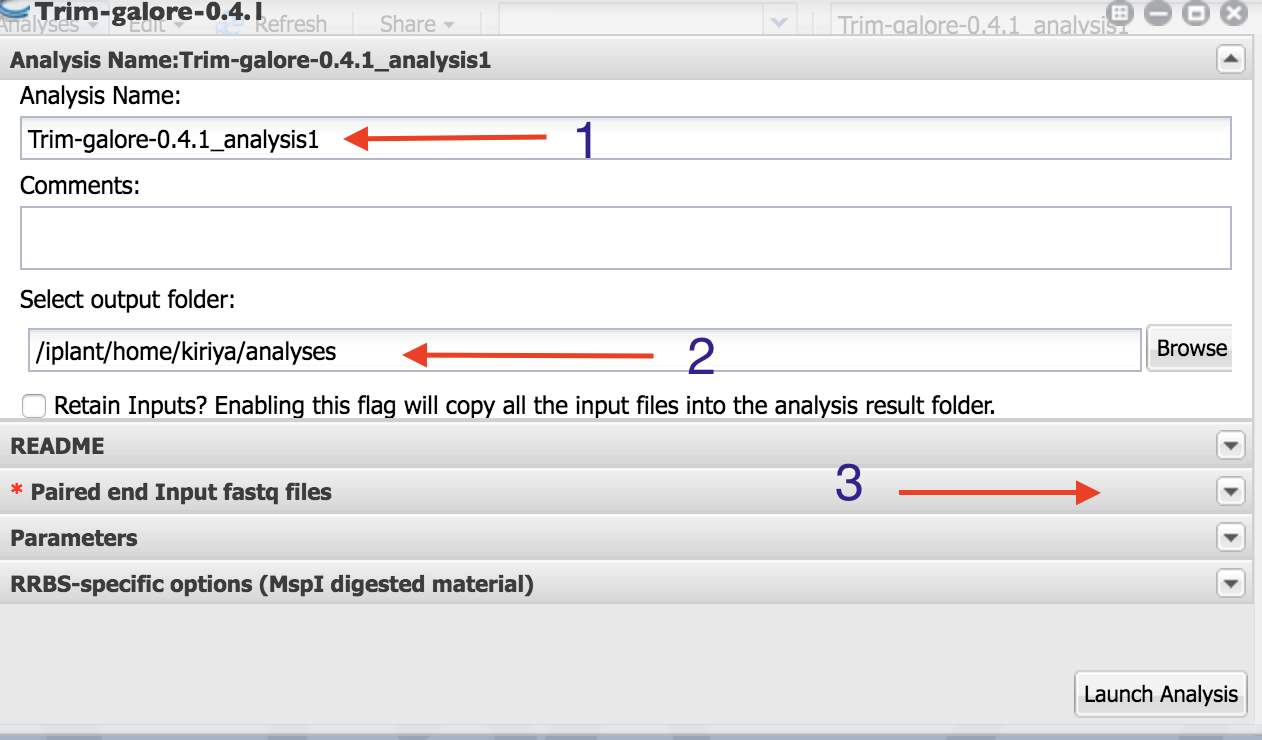
Trim Galore In R . this script runs the trim galore! This function calls trim_galore using system2(), and is only designed to handle standard. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. In this mode, trim galore identifies. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Tool and requires installation of both cutadapt and trim galore!
from dxoooycal.blob.core.windows.net
this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. In this mode, trim galore identifies. This function calls trim_galore using system2(), and is only designed to handle standard. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Tool and requires installation of both cutadapt and trim galore!
Trim Galore Cutadapt at Christine Gonzales blog
Trim Galore In R this script runs the trim galore! this script runs the trim galore! In this mode, trim galore identifies. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Tool and requires installation of both cutadapt and trim galore! This function calls trim_galore using system2(), and is only designed to handle standard. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data.
From www.researchgate.net
Feature comparison of FastQC, Trimmomatic, Cutadapt, and AfterQC Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. This function calls trim_galore using system2(), and is only designed to handle standard. In this mode, trim galore identifies. Tool and requires installation of both cutadapt and trim galore! Is a wrapper script to automate quality and adapter trimming as well as. Trim Galore In R.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore In R Tool and requires installation of both cutadapt and trim galore! trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming. Trim Galore In R.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore In R this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Tool and requires installation of both cutadapt and trim galore! trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. This function calls. Trim Galore In R.
From www.researchgate.net
Passo a passo para execução da ferramenta Trim Galore! na plataforma Trim Galore In R This function calls trim_galore using system2(), and is only designed to handle standard. In this mode, trim galore identifies. Tool and requires installation of both cutadapt and trim galore! this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate. Trim Galore In R.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore In R trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Tool and requires installation of both cutadapt and trim galore! this script runs the trim galore! This function calls trim_galore using system2(), and is only designed to handle standard. In this mode, trim galore identifies. Is a. Trim Galore In R.
From nf-co.re
modules/trimgalore Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. In this mode, trim galore identifies. Tool and requires installation of both cutadapt and trim galore! This. Trim Galore In R.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore In R In this mode, trim galore identifies. Tool and requires installation of both cutadapt and trim galore! This function calls trim_galore using system2(), and is only designed to handle standard. this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. trim galore is a bioinformatics. Trim Galore In R.
From zymo-research.github.io
How to interpret the bulk RNAseq report pipelineresources Trim Galore In R In this mode, trim galore identifies. this script runs the trim galore! trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Tool and requires installation of both cutadapt and trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control,. Trim Galore In R.
From zymo-research.github.io
How to interpret the small RNAseq report pipelineresources Trim Galore In R This function calls trim_galore using system2(), and is only designed to handle standard. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Tool and requires installation of both cutadapt. Trim Galore In R.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Tool and. Trim Galore In R.
From blog.csdn.net
保姆级有参考RNAseq分析基本流程1(表达矩阵)_rnaseq比对定量后如何获得基因表达矩阵CSDN博客 Trim Galore In R Tool and requires installation of both cutadapt and trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. this script runs the trim galore! trim galore is a bioinformatics tool. Trim Galore In R.
From dxoooycal.blob.core.windows.net
Trim Galore Cutadapt at Christine Gonzales blog Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. This function calls trim_galore using system2(), and is only designed to handle standard. In this mode, trim galore identifies. this script runs the. Trim Galore In R.
From gensoft.pasteur.fr
image Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. this script runs the trim galore! In this mode, trim galore identifies. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. This function calls trim_galore using system2(), and. Trim Galore In R.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore In R Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter. Trim Galore In R.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore In R This function calls trim_galore using system2(), and is only designed to handle standard. this script runs the trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data.. Trim Galore In R.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore In R This function calls trim_galore using system2(), and is only designed to handle standard. this script runs the trim galore! Tool and requires installation of both cutadapt and trim galore! Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Is a wrapper script to automate quality and adapter trimming as well. Trim Galore In R.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore In R trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. In this mode, trim galore identifies. This function calls trim_galore using system2(), and is only designed to handle standard. Tool. Trim Galore In R.
From www.youtube.com
[실습] Trim Galore를 사용한 FASTQ 파일 전처리 (1)* YouTube Trim Galore In R In this mode, trim galore identifies. trim galore is a bioinformatics tool designed to simplify the process of quality control and adapter trimming of raw sequencing data. this script runs the trim galore! This function calls trim_galore using system2(), and is only designed to handle standard. Is a wrapper script to automate quality and adapter trimming as well. Trim Galore In R.